Semnet: Semantic networks from embeddings
I’m happy to introduce Semnet, a small Python library which efficiently constructs graph structures from embeddings.
The name “Semnet” derives from semantic network, as it was initially designed for an NLP use-case, but it will work well with any form of embedded document (e.g., images, audio, even or graphs).
In this post, I’ll quickly run over some of the features and use cases, discuss why Semnet exists and the benefits of graph structures in natural language processing and beyond.
If you want to know more, I have another hands-on post providing examples of Semnet in action. You can also read the docs and check out the repository on Github.
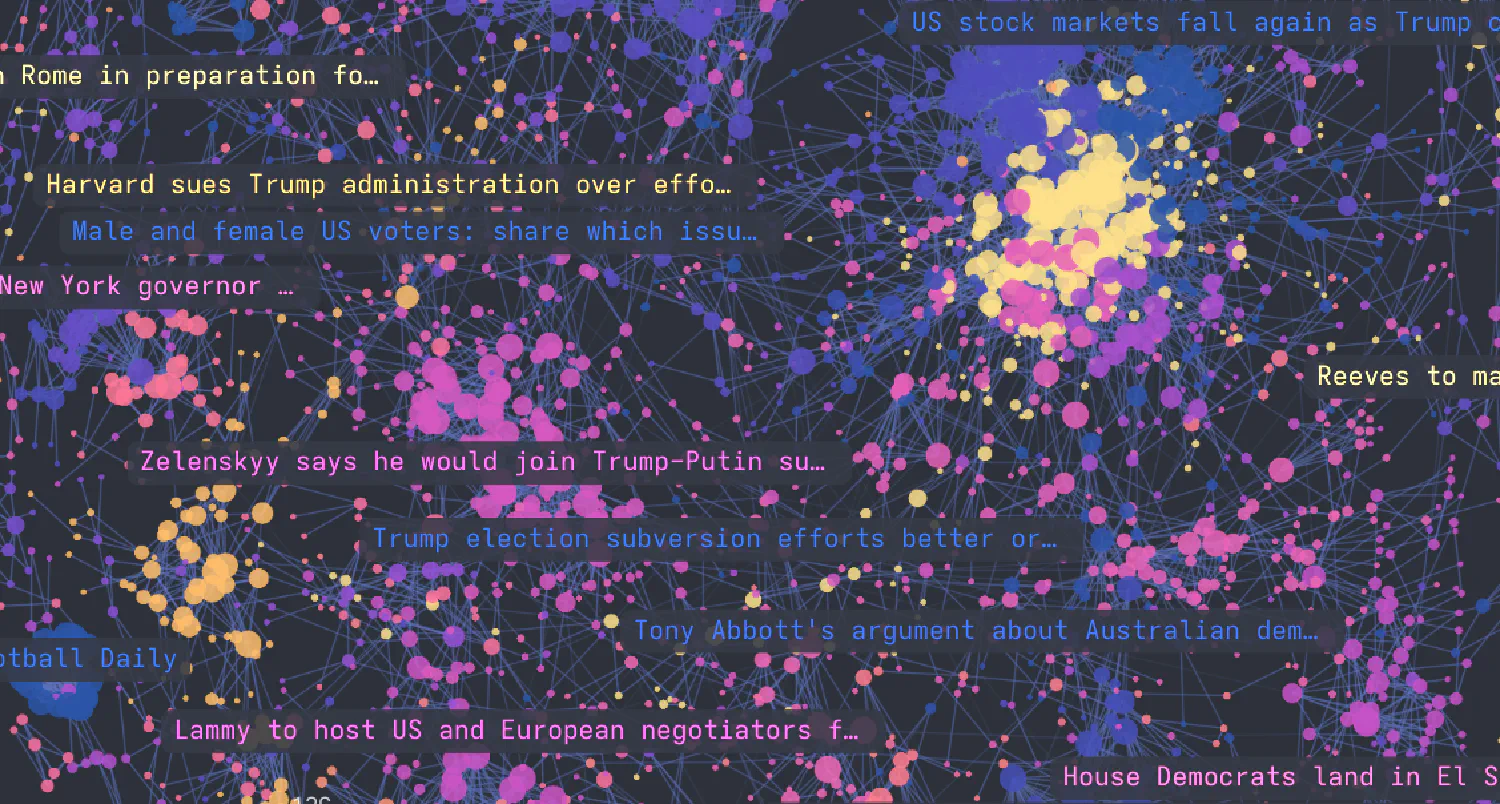
If you’d just like to see something cool you can do with Semnet, click on the image below for an interactive visualisation of a network of quotes.
 Graph built with Semnet and visualised in Cosmograph. Data: m-ric via Hugging Face.
Graph built with Semnet and visualised in Cosmograph. Data: m-ric via Hugging Face.
Features
Semnet is a relatively small library, which does just one thing: efficiently construct network structures from embeddings.
Its key features are:
- Rapid conversion of large embedding collections to graph format, with nodes as documents and edges as document similarity.
- It’s fast and memory efficient: Semnet uses Annoy under the hood to perform efficient pair-wise distance calculations, allowing for million-node networks to be constructed in minutes on consumer hardware.
- Graphs are returned as NetworkX objects, opening up a wide range of algorithms for your project.
- Pass arbitrary metadata during graph construction or update it later in NetworkX.
- Control graph construction by setting distance measures, similarity cut-offs, and limits on outbound edges per node.
- Easily convert to pandas for downstream use.
Use cases
Semnet may be used for:
- Graph algorithms: enrich your data with communities, centrality and much more for use in NLP, search, RAG and context engineering.
- Deduplication: remove duplicate records (e.g., “Donald Trump”, “Donald J. Trump) from datasets.
- Exploratory data analysis and visualisation, Cosmograph works brilliantly for large corpora.
Quick start
You can easily install Semnet with pip.
pip install semnet
All you need to start building your network is a set of embeddings and (optionally) some labels.
from semnet import SemanticNetwork, to_pandas
from sentence_transformers import SentenceTransformer
# Your documents
docs = [
"The cat sat on the mat",
"A cat was sitting on a mat",
"The dog ran in the park",
"I love Python",
"Python is a great programming language",
]
# Generate embeddings (use any embedding provider)
embedding_model = SentenceTransformer("BAAI/bge-base-en-v1.5")
embeddings = embedding_model.encode(docs)
# Create and configure semantic network
sem = SemanticNetwork(thresh=0.3, distance="angular")
# Build the semantic graph from your embeddings
G = sem.fit_transform(embeddings, labels=docs)
# Analyze the graph
print(f"Nodes: {G.number_of_nodes()}")
print(f"Edges: {G.number_of_edges()}")
# Export to pandas
nodes_df, edges_df = to_pandas(G)
What problem does Semnet solve?
Graph construction entails finding pairwise relationships (edges) between entities (nodes) in a dataset.
For large corpora, scaling problems rapidly become apparent as the number of possible pairs in a set scales quadratically.
$$pairs = \frac{n(n-1)}{2}$$
Naive approach
If we were to naively attempt to construct a graph from a modestly-sized set of documents we encounter problems early on with modestly-sized corpora. For example, building a graph from 10,000 documents would entail operations across 50 million pairs, for 100,000 it’s around 5 billion!
Iterating over each pair is of course very slow. Faster approaches exist via scikit-learn, but here we run into a larger problem: it’s memory intensive:
from sklearn.metrics import DistanceMetric
import numpy as np
dist = DistanceMetric.get_metric("euclidean")
# Generate 10_000 random embeddings
embeddings = np.random.rand(100_000, 768)
dist_scores = dist.pairwise(embeddings)
>> MemoryError: Unable to allocate 74.5 GiB for an array
with shape (100000, 100000) and data type float64
With Semnet
Semnet solves the scaling problem using Approximate Nearest Neighbours search with Annoy.
Instead of making comparisons between each document in the corpus, Semnet indexes the embeddings, iterates over each one, and returns a top_k best matches from within their neighbourhood.
Trying this again on the same rig with Semnet.
from semnet import SemanticNetwork
# Kick off timer again
start_time = time.time()
# Make 100,000 copies this time
embeddings = np.random.rand(100_000, 768)
# Build semantic network
semnet = SemanticNetwork(thresh=0.4, top_k=5)
G = semnet.fit_transform(embeddings)
# Close off the timer
end_time = time.time()
print(f"Processing time: {end_time - start_time:.2f} seconds")
>> Processing time: 24.26 seconds
We’re not only able to process all the embeddings without crashing our kernel, but it’s done in under 30 seconds.
Why use graph structures?
By opening up the NetworkX API to embedded documents, Semnet provides a new suite of tools and metrics for workflows in domains such as NLP, RAG, search and context engineering.
For most use cases, Semnet will work best as a complement to traditional spatial workflows, rather than as a replacement. Its power lies in encoding information about relationships between data points, which can be used as features in downstream tasks.
Approaches will vary depending on your use case, but benefits include:
- Accessing network structures like paths, local neighbourhoods and subgraphs
- Using centrality measures and communities that capture relationships and structure
- Making some very pretty visualisations
Want to know more?
- Head over to the examples blog
- Read the docs
- Check out the repository on Github

